See the preprint for a thorough discussion or to cite this resource:
ExPheWas: a browser for gene-based pheWAS associations.
Legault, M.-A., Lemieux Perreault, L.-P., & Dubé, M.-P. (2021). medRxiv. https://doi.org/10.1101/2021.03.17.21253824
Overview
The ExPheWas browser showcases the results of a phenome-wide association study (PheWAS) using an approach that models the joint effect of variants at protein coding genes. In the original release, we tested the association between 19,114 genes on human autosomes and 1,210 phenotypes available from the UK Biobank.
Briefly, we used a principal component analysis of common variant genotypes (PCA) to reduce the effective number of variables needed to represent the genotypes. Association testing is then done by assessing the improvement in goodness of fit from including the gene based principal components to a regression model. The experimental setup is described in more details in the figure below:
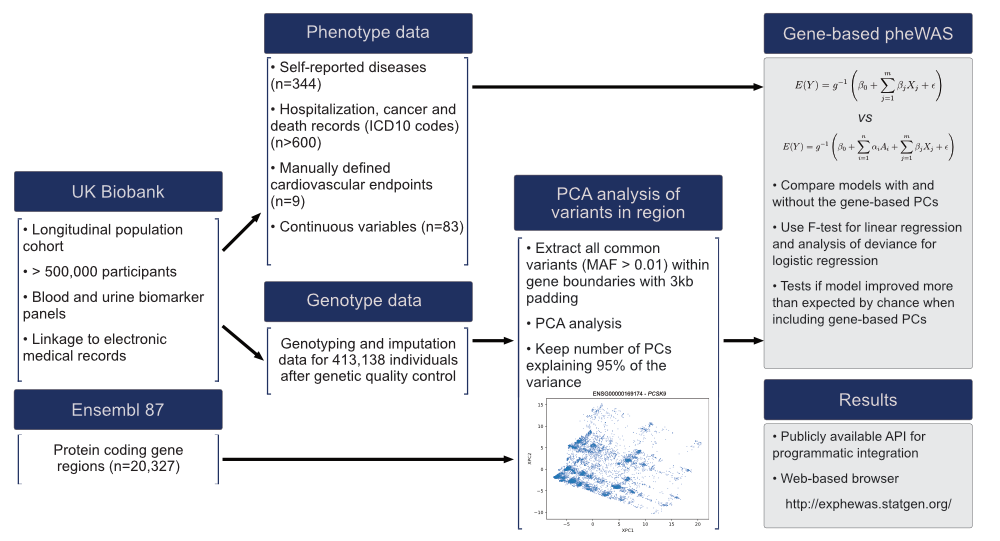
PheWAS results can then be browsed by gene or by phenotype. The pages are mostly self-explanatory, but key features are illustrated below:
Support
For technical assistance or feature requests, we recommend you open a ticket on the project's Github page. For other inquiries, you may contact the corresponding author of the manuscript.